Projects
Assembly of retrovirusesLate in the retroviral life cycle, immature viral particles are assembled from polyprotein precursors, translational products of the gag, gag-pol or gag-pro, and gag-pro-pol genes, and two molecules of genomic RNA. Assembly can take place either in the cytoplasm of infected cells (so called D-type viruses, i.e. M-PMV and MMTV) or at the plasma membrane, concomitant with the budding process (so called C-type viruses, e.g. HIV). We have used both morphologic types of retroviruses for assembly process analysis. In collaboration with Prof. Ruml's laboratory at ICT Prague, we have shown that M-PMV Gag polyprotein can assemble into procapsids in prokaryotic cells and in an in vitro system. Using both these systems, we have identified the minimal domain of M-PMV Gag ( ProCANC) necessary for the assembly of immature particles. Current research is focused on identifying compounds that efficiently inhibit in vitro assembly of HIV CA and CANC proteins and, in cooperation with foreign laboratories, we will evaluate their influence on virus assembly in infected cells. We are also investigating the structure of M-PMV immature capsids and the role of nucleic acids in the assembly of M-PMV. Additionally, studies focused on identifying cellular proteins involved in intracellular transport of M-PMV Gag polyproteins to the site of assembly are underway. In collaboration with the NMR laboratory at ICT Prague, we implement structural analyses of viral proteins participating in the assembly process. Furthermore, we are also performing in vitro and in vivo experiments aiming to define an internal scaffold domain that permits intracytoplasmic assembly of the Mouse mammary tumor virus. |
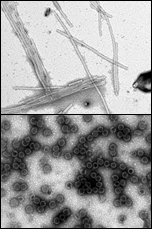
|
Maturation of retrovirusesThe retroviral proteins are synthesized as Gag polyprotein precursors, which are processed by virus encoded protease (PR) during or shortly after budding. This proteolytic cleavage triggers the particle re-arrangement into mature, fully infectious particles. Our main goal is to investigate a mechanism of activation of PR, i.e. the initiation of cleavage of the PR domain from polyprotein precursors and the formation of the active dimeric form of PR. Because the processes of capsid assembly and maturation in Mason-Pfizer monkey virus are temporally and spatially distinct from one another, we are able to use M-PMV as a model for these studies. M-PMV protease contains unique C-terminal extension sequences with glycine-rich motifs, known as G-patch, that are removed in vitro as well as in vivo by autoproteolytic processing to yield truncated active forms of PR. NMR structural analyses and biochemical experiments have confirmed that this domain is not necessary for the formation of fully folded active protease. To investigate the role of the G-patch domain in virus replication, we use several different approaches including the yeast two hybrid system to screen for interacting host proteins, immunofluorescent microscopy for analysis of G-patch trafficking, and mutational analysis of functionally important sequences in M-PMV protease followed by an assessment of virus processing, infectivity, and morphology. |

|
Retroviral integrationThe process of establishing the proviral DNA in the host chromosome involves the transport of a complex of associated viral proteins, cellular proteins, and nucleic acids (termed preintegration complex, PIC) from cytoplasm into the nucleus where integration is then catalyzed by a virus encoded integrase. We are working towards identifying the M-PMV PIC constituents by immunochemical detection and MS analysis. Developing and testing HIV integrase inhibitors is also currently performed. |
|
Role of secreted aspartic proteases (Saps) in pathogenic yeast metabolism and in host-pathogen interactionsCandida species are ubiquitous human fungal pathogens that can cause life threatening infections in immunocompromised patients. The secretion of proteases is one of the virulence factors of pathogenic Candida spp. Together with lipases and phospholipases they facilitate the penetration of host surfaces. Therefore the specific inhibitors of Saps are considered a possible antimycotic drug target. Our current work is focused on: obtaining more detailed knowledge of the structure of Saps in C. albicans, C. tropicalis, and C. parapsilosis, mapping substrate specificities, sap gene regulation, analysis of intracellular Sap precursor trafficking, activation, and secretion of mature proteases. |

|
Functional analysis of insect desaturasesDesaturases participate in pheromone production in various insect species. Desaturase introduces a double bond into the aliphatic chain of the fatty acids with variable specificity. The pheromone blend produced by our model organism Manduca sexta is unusually complex and contains mono-, di- and trienes. To investigate the mechanism of pheromone synthesis and the role of different types of desaturases in the production of pheromones, we have constructed cDNA libraries from mRNA isolated from pheromone glands and fatty bodies and have constructed the genes encoding their respective desaturases. Together with gene analysis, we are performing a functional expression of the insect desaturases in Saccharomyces cerevisiae followed by GC-MS analysis of fatty acids methyl esters and their derivatized products, which help us identify the class of isolated desaturases. This project is being investigated in close cooperation with Dr. Svatoš from the Max Planck Institute for Chemical Ecology in Jena. We are also collaborating with Dr. Valterová's group at IOCB on the identification of desaturases in the labial gland of male Bombus lucorum. |

|