Projects
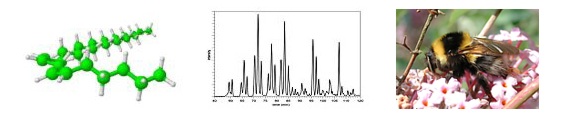
Analysis of complex lipid mixturesLipids are very important constituents of cellular organisms. They have many functions including storage of metabolic energy, signaling, membrane formation, and surface protection. Natural lipids usually form very complex mixtures. Their analysis is quite challenging and plays an indispensable role in lipidomics, which studies complex relationships between lipids in the living organisms. Our group develops new analytical methods for their characterization using advanced separation and mass spectrometric techniques. Samples are isolated from various organisms, particularly from insect and plants. Characterizing triacylglycerols in insect fat body, particularly in bumblebees, is one of our projects. The previous studies were almost exclusively limited to determination of fatty acids using transesterification and GC analysis. Important information about bonding of fatty acids to the glycerol backbone is lost by this procedure. We optimize high-performance liquid chromatography - atmospheric pressure chemical ionization mass spectrometry for thorough characterization of individual isomers. Two separation systems, non-aqueous reversed-phase and silver ion (cation exchange resin in silver (I) ionic form) are used to achieve the best possible chromatographic resolution. Individual components are then characterized from their mass spectra. Spectra interpretation is facilitated by software tools developed in our group (see bellow). |
|
|
Matrix-assisted laser desorption/ionization (MALDI) is employed for rapid characterizing of lipid mixtures. MALDI mass spectra do not provide as much as information as HPLC/MS, however, the data are useful e.g. for statistical evaluation of the sample, or for chemotaxonomic purposes. Various lipid classes are being analyzed, including glycerolipids, glycolipids, wax esters etc. Hydrocarbons are perhaps the most difficult compounds to characterize by MALDI. We study them because of their interesting functions on the insect or plant cuticle taking part in a variety of plant-insect interactions. Hydrocarbons are usually analyzed by GC/MS. Both GC and electron ionization MS have, however, limitations for high molecular weight compounds and GC/MS thus detecs only part of the mixture components. MALDI analysis of hydrocarbons is advantageous because extents molecular weights range considerably. Hydrocarbon ionization under MALDI conditions is not, however, straightforward, as they lack polar group prone to protonation or cation attachment. We showed, that hydrocarbons are ionized by lithium when lithium 2,5?dihydroxybenzoate matrix is used. Using this method, hydrocarbons of various insect species were characterized. Several previously unknown hydrocarbons were discovered. However, their structure and biological function still needs to be disclosed.
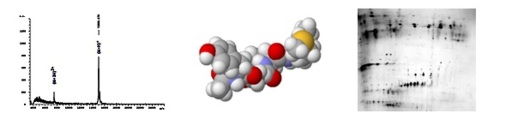
Proteins and peptides structure analysisMass spectrometry group participates in number of projects where structure of peptides and proteins is an essential knowledge. |
|
|
Mass spectrometers are used for identification of biologically active peptides, particularly with antimicrobial effects. Tandem mass spectrometry is used for “de-novo” sequencing. Biomarkers of serious diseases are searched and identified in complex protein samples. Peptide mass fingerprinting as well as peptide sequencing and database searching are among the most frequently used tools. To study expression level of proteins of interest, various quantitation methods are employed. Matrix-assisted laser desorption/ionization is also used for rapid identification of bacteria strains and building up corresponding databases.
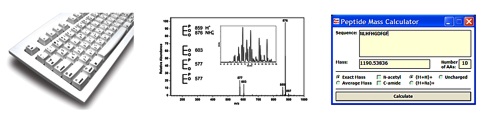
Computer-assisted mass spectra interpretationWhen mass spectra library search is not available, or do not provide reliable results, manual spectra interpretation is performed. Manual interpretation is usually time consuming and often a nightmare for a person who is not an expert in mass spectrometry. In many cases software tools can be developed to assist with the spectra interpretation. The use of such tools is particularly advantageous when many similar mass spectra of related compounds have to be processed. Our group develops software tools to assist mass spectra interpretation. |
|
|
Our approach is demonstrated on APCI mass spectra of triacylglycerols. Natural triacylglycerols exist as complex mixtures of isomers in which fatty acid chains vary from one molecule to the next. APCI spectra are relatively simple showing molecular adduct(s) and fragment ions in the positive ion mode. Interpretation is usually straightforward, consisting of the determination of fatty acid neutral losses. Taking into account that tens to hundreds of isomers is usually present in the mixtures, complete interpretation of HPLC/MS data is very time-consuming, which requires much more time than the sample analysis. Therefore, software named TriglyAPCI was developed to greatly speed up the interpretation procedure and also preclude possible mistakes. The algorithm can be adapted to interpret also mass spectra of other lipids where the FAs in the chains vary. In the other project GC/EI-MS data are used to locate branching position(s) in cuticular hydrocarbons. |