JANA JERSÁKOVÁ, PAVEL TRÁVNÍČEK, BARBORA KUBÁTOVÁ, JANA KREJCÍKOVÁ, TOMÁŠ URFUS, ZHONG-JIAN LIU, ANTHONY LAMB, JAN PONERT, KATHARINA SCHULTE, VLADISLAV ČURN, JAN VRÁNA, ILIA J. LEITCH and JAN SUDA
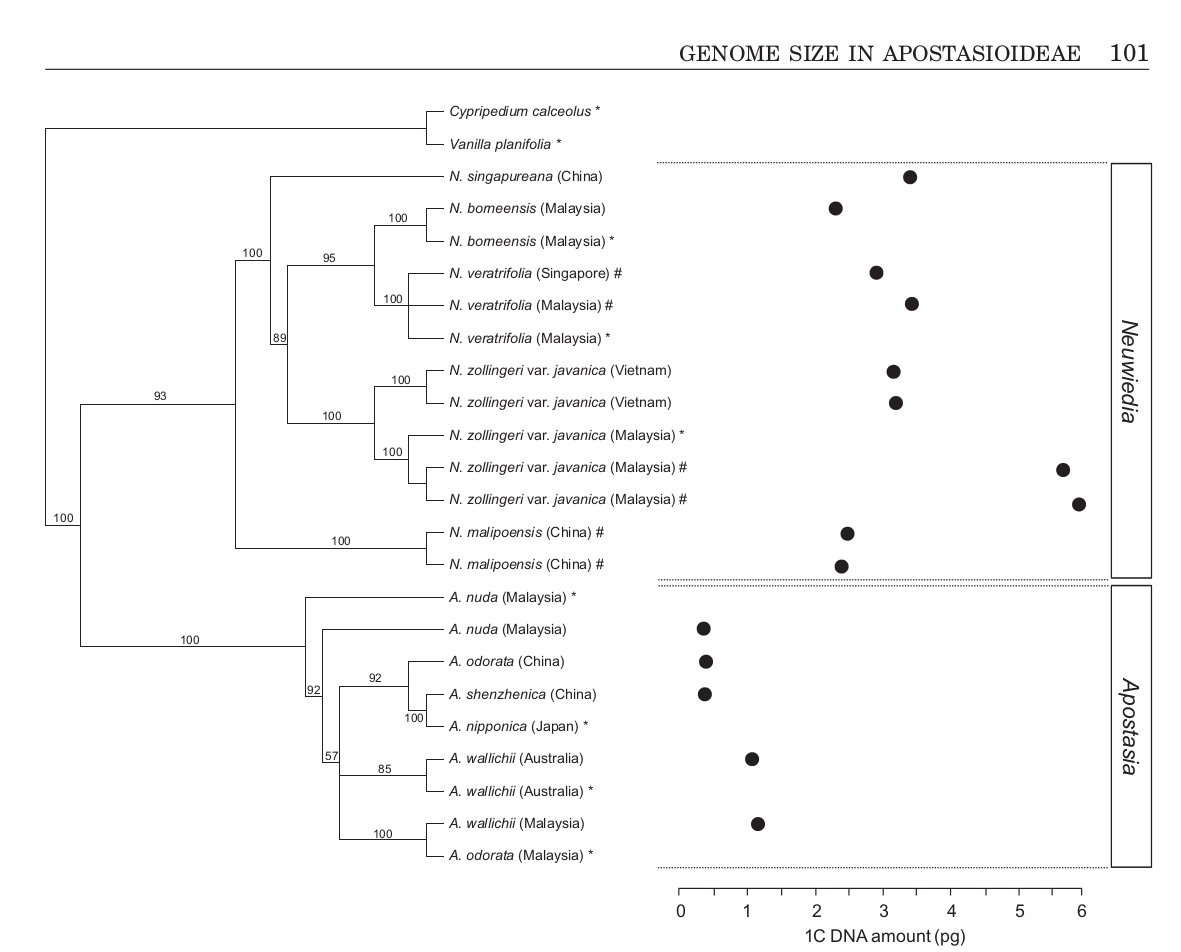
With more than 160-fold variation, Orchidaceae are currently the most diverse angiosperm family with respect to the amount of nuclear DNA. This study provides first genome size estimates for approximately 50% of species currently recognized in subfamily Apostasioideae, which is sister to the other four orchid subfamilies. The estimated 1C-values range from 0.38 pg in Apostasia nuda to 5.96 pg in Neuwiedia zollingeri var. javanica, a nearly 16-fold range. The two genera show non-overlapping genome sizes, with those in Apostasia being distinctly smaller than those in Neuwiedia. In fact, most Apostasia spp. are at the lower end of the range of orchid C-values.
Observed discontinuities in DNA amounts in genera most probably reflect interspecific variation in ploidy. In addition to ploidy heterogeneity in N. zollingeri var. javanica, intraspecific variation in genome size (up to 17.7%) was also detected in some species; this can be plausibly related to the incidence of different geographical variants
or unrecognized taxonomic heterogeneity. The AT content varied from 62.6 to 66.0%, which is in the upper range recorded for angiosperms. The genome size data obtained in this study fill a major phylogenetic gap in Orchidaceae and show that (very) small genomes prevail in subfamily Apostasioideae.