Chromatin structure (e.g. heterochromatin content) may be evaluated by flow cytometric analysis of nuclei stained by fluorochromes which bind to DNA by different modes (e.g. intercalation / binding to major or minor groove of DNA molecule).
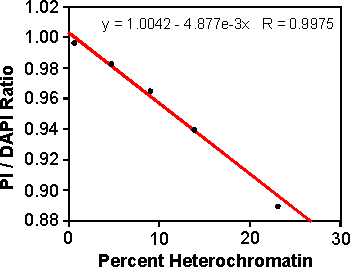
Figure 26. Linear regression of Propidium Iodide (PI) / DAPI fluorescence ratio versus percentage heterochromatin in maize lines. (Rayburn et al., 1992)
Structural chromosome aberrations in dividing cells lead to unequal distribution of DNA into daughter cells. This is reflected by the increase in variation of nuclear DNA content (increase of coefficient of variation of G1 and G2 peaks). In addition, higher doses may result in changes of cell cycle kinetics.

Base composition of nuclear DNA can be estimated by separate analyses using fluorochromes showing preference to AT- or GC- rich regions of DNA relative to a known standard.

If sex chromosomes differ sufficiently in size (and hence in DNA content), or in DNA base composition, flow cytometric analysis of isolated nuclei may be used for early screening of sex in non-flowering individuals.

In allopolyploid species whose basic genomes are known to be of different size, it is possible to use flow cytometric measurement of nuclear DNA content for estimation of their genome composition.

Comparison of relative positions of G1 peaks corresponding to the sample nuclei and the nuclei isolated from a plant with known DNA content, respectively, permit accurate determination of the "unknown" DNA content:


Since the nuclear DNA content reflects the position of a cell within a cell cycle, flow cytometric analysis of nuclear DNA content is suitable for cell cycle analysis. In order to determine the fraction of cell population in the G1, S and G2 phases, a DNA content distribution must be deconvolved (most conveniently using a dedicated computer software).

In some plant species (e.g. in most of angiosperms), cell differentiation may be accompanied by endopolyploidization either viaendomitosis or endoreduplication. Thus, in addition to diploid cells in G1-, S-, or G2- phase of the cell cycle, differentiated plant tissues may contain endopolyploid cells whose DNA content ranges from 4C to 128C or even more.

Because the nuclear DNA content of G1 nucleus reflects the ploidy of a cell, estimation of DNA content is frequently used for ploidy determination.
Table 2. Relation between the ploidy and DNA content of G1 phase nuclei
1a. Chop a small amount of plant material (typically 20 mg) with a new razor blade or a sharp scalpel in 1ml of ice-cold LB01 in a petri dish. It is preferable to include a DNA fluorochrome (DAPI or propidium iodide) in the buffer prior to chopping. Alternatively, this compound may be added immediately after the filtration (step 2). The stains are used in the following concentrations: DAPI, 2 µg/ml; propidium iodide, 50 µg/ml + RNase, 50 µg/ml.