2010
|
DNA decorated by aldehydes for attachment of other moleculesA novel simple and efficient methodology for attachment of other molecules to DNA (bioconjugation) was developed. It consists in synthesis of DNA bearing very reactive chemical functional groups that can be readily (in one step) linked to diverse other molecules e.g. for studying of molecular mechanism of important biological processes or for labeling of DNA by color or electroactive markers. This methodology is much shorter, simpler and easier than existing methods of preparation of DNA conjugates and thus it has a promising potential for a broad range of applications in interdisciplinary area between chemistry and biology.
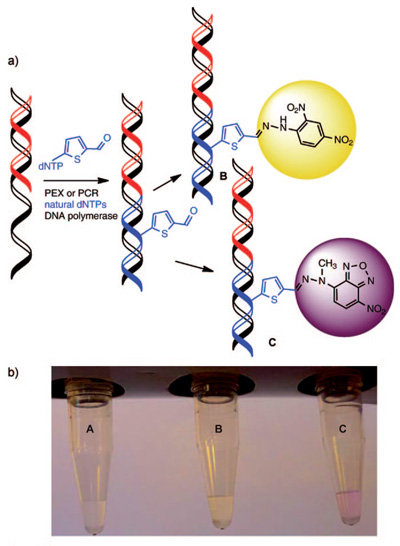 Key intermediate is a DNA bearing very reactive chemical functional groups (aldehydes) as molecular "clips". The methodology of its synthesis is very straightforward and consists of only two steps. The first step is chemical synthesis of modified nucleoside triphosphates and the second one is enzymatic polymerase catalyzed synthesis of DNA from these building blocks. In this way, one can prepare both short sequences containing one or several aldehyde "clips" and very long DNA containing hundreds of such groups. The aldehyde groups readily react with a number of reagents to attach virtually any other molecule. This principle was shown on the formation of colored compounds (hydrazones) used for staining of DNA to yellow or pink color (see Figure). Now other reactions of the modified DNA are studied in order to attach important biomolecules (peptides, proteins etc.). This methodology may find a wide range of applications not only in preparation of diverse bioconjugates of DNA but also in material science or nanotechnology where the DNA could serve as an easily programmable and renewable scaffold for attachment of useful chemical molecules of functional groups.
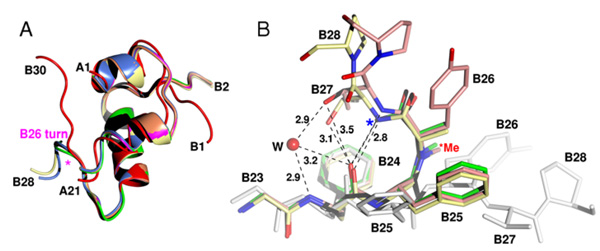
Modified insulins help to elucidate the mechanism of insulin actionInsulin is a key protein hormone that regulates blood glucose levels and, thus, has widespread impact on lipid and protein metabolism. Insulin resistance or failure to synthesize insulin de novo leads to diabetes mellitus. In 2009, at least 245 million people worldwide had diabetes. Insulin action is manifested through binding of its monomeric form to the specific membrane insulin receptor. Despite substantial effort of many laboratories, the structure of the insulin-receptor complex is not known and our knowledge about structural behavior of insulin is based upon inactive, multimeric, storage-like states. It is widely acknowledged that insulin must undergo structural changes upon binding to the receptor. We prepared a series of highly active insulin analogues with modifications at the B26 position. X-ray structural analysis revealed unique 3-D structures. The main feature of these new insulin structures is a β-turn at the C-terminus of the B-chain of insulin ("B26 turn", left Figure) associated with a trans to cis isomerisation of the B25-B26 peptide bond (right Figure). The resultant conformational changes unmask previously buried amino acids that are implicated in receptor binding. We believe that the structures of these novel insulin analogues are very similar to the active form of the hormone. Our results represent a milestone in the study of insulin interaction with the receptor. Modified insulins resembling the active form of the hormone may be the starting point for the development of new insulin analogues or insulin mimetics for nasal, pulmonal or oral treatment of diabetes.
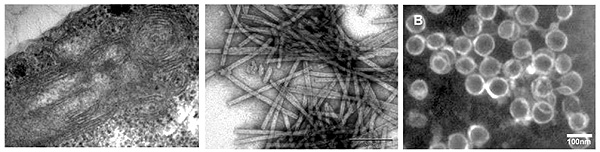
Structure motifs of Gag polyproteins important for assembly of retroviral particlesAssembly of viral particles in the host cells represents one of the key steps of the replication cycle of retroviruses. The viral proteins are synthesized in a form of precursors (Gag polyproteins) and are transported to place of assembly in the host cells where together with genomic RNA form immature viral particles. Detailed knowledge of mechanism and identification of structure motifs influencing the assembly is important for design of novel type of inhibitors, which block the protein-protein and /or protein-nucleic acid interactions during the assembly process. We have used Mason-Pfizer monkey virus (M-PMV) and Mouse mammary tumor virus, (MMTV) as models. Analysis of the large series of the structural protein mutants using the in vitro assembly system that we developed in our laboratories, in combination with experiments with proviral constructs in the host cells, has enabled us to identify domains and structural motifs essential for the assembly of immature and mature particles. The results were published in Journal of Virology. We have also developed a rapid, simple and high-throughput method for monitoring the assembly of HIV-1 immature particles suitable for testing libraries of compounds inhibiting this process. This method is based on interaction of fluorescently labeled oligonucleotide (ON) that specifically binds to the retroviral structural proteins and initiates the in vitro assembly of HIV immature particles. The method was patented.
|