Projects
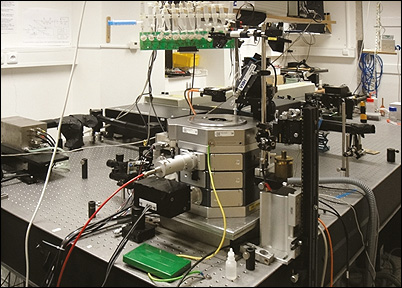
Improving the implementation of 2-photon polarization microscopy & related techniquesWe are working on combining two-photon polarization microscopy (2PPM) with techniques of superresolution, non-scanning two-photon microscopy, holographic techniques, light sheet microscopy and others, in order to reach spatial and temporal resolutions and sensitivity needed to observe the various molecular processes that are of interest to us and our collaborators. We are trying to explore fast biological processes, such as the action potential in in neurons, or light-activated events, which we would like to be able to observe on a 100 ns time scale. We are exploring optical probes other than GFP-like fluorescent proteins.
 |
|
Development of two-photon polarization microscopy into a tool of structural biologyThe 2PPM technique allows sensitive observations of changes in protein conformation in living cells and organisms. We are continuously working on expanding the capabilities of the technique in this area, so that we could use 2PPM to make quantitative insights into the structure of membrane proteins in living cells. In order to establish the ability to make such structural insights, we have turned to highly controlled, simple biophysical models (synthetic lipid vesicles, small molecule dyes; Timr Š. et al.: J. Phys. Chem. B 2014), and we have been gradually applying the technique to more complex and biologically relevant systems. To guide our experiments and to verify (or not) our results, we rely on quantum mechanical (QM) and molecular dynamics (MD) simulations, carried out in close collaboration with the computational chemistry group of Pavel Jungwirth (IOCB). In protein structure studies we collaborate with the group of Antoine Royant at the ESRF in Grenoble.
 |
|
Development of genetically encoded optical probesTwo-photon polarization microscopy allows development of novel genetically encoded optical probes of molecular processes taking place in living cells and organisms. In fact, many existing fluorescently labeled constructs can, without any modification, serve as probes of protein-protein interactions or conformational changes. Probes based on 2PPM have important advantages over other techniques (such as FRET): they only need a single fluorescent moiety, and allow quantitative structural insights. We are mainly working on development of novel genetically encoded sensors of cell membrane voltage (collaboration with Lawrence B. Cohen at Yale University, Han Z. et al, PLoS One 2014), and development of sensors of G-protein activation (several collaborations). However, other processes (EGF signaling, neurotransmitter transport, calcium signaling) are also of interest to us. Development of new probes is beginning to be guided by quantitative insights into protein structure that 2PPM allows.
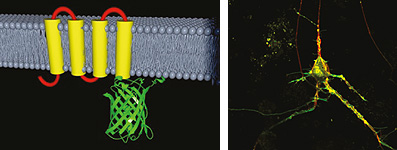 |
|
Applications of 2PPM to answering biological questionsWe are working on elucidating various aspects of molecular mechanisms of GPCR and G-protein signaling (Bondar A. & Lazar J.: J. Biol. Chem. 2014): issues of GPCR dimerization, GPCR - G-protein pre-coupling, specificity of GPCR - G-protein interactions, G-protein rearrangement/dissociation and others. We are interested in molecular mechanisms of insect and mammalian olfaction and pheromone perception. Our 2PPM-based probes allow observing molecular systems closer to their natural state than what is allowed by other probes. We are interested in development of high throughput/high content screens useful in pharmaceutical research.
 |
|