2008
|
N6-Methyl-AMP aminohydrolase - a key enzyme of N6-substituted purine acyclic nucleoside phosphonates intracellular activationIt was shown that this so far unknown enzyme catalyzes the hydrolytic deamination of natural substrates N6-meAMP, N6,N6-dimethyl-AMP and N6-medAMP to IMP and/or dIMP, respectively. Instead of its natural metabolic function, this enzyme efficiently converts N6-substituted prodrugs of acyclic nucleoside phosphonates to the active guanine analogs, potential antineoplastic and antiviral agents. The deamination of N6-cyclopropyl-2,6-diamino-9-[2-(phosphonomethoxy)ethyl]purine (cyprPMEDAP), an intermediate intracellular metabolite of antileukemic agent GS-9219 to the corresponding active nucleotide analog 9-[2-(phosphonomethoxy)ethyl]guanine (PMEG) is an example.
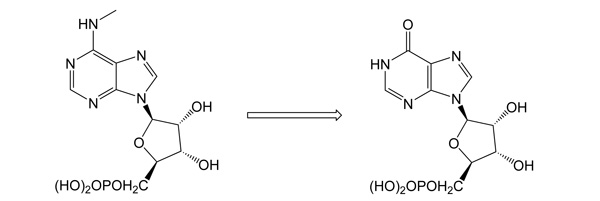
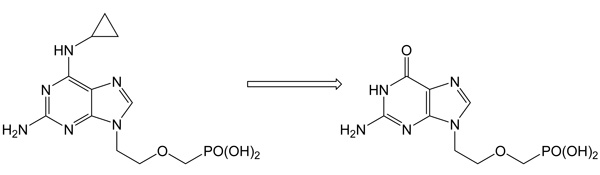
Schinkmanová M., Votruba I., Shibata R., Han B., Liu X., Cihlar T., Holý A.:
Protein association in salt solutions: Molecular understanding of the Hofmeister series120 years after Franz Hofmeister (at the Pharmacological Institute in Prague) ordered salt ions according to their ability to salt-out proteins, the present molecular dynamics simulations provide a key to the molecular understanding of the lyotropic (Hofmeister) series. Simulating protein-protein interactions in aqueous alkali halide solutions we showed that this ion ordering is due to a multitude of different effects rather than due to a single one. Indeed, for the model case of association of lysozyme molecules we clearly observed at least two such effects. These are the direct interaction of aqueous anions with positively charged amino acid residues and the affinity of these anions for hydrophobic patches at protein surfaces. While the former interactions are stronger for chloride than for iodide, the opposite is true for the latter effect. The protein-protein interaction in a specific salt solution is then a result of a subtle balance between these (and also other) forces. In the present case, the hydrophobic effect of iodide wins over the ion-pairing effect of chloride, which results in a stronger lysozyme-lysozyme association in aqueous NaI than NaCl. The present results, which are supported by experiments, have implications not only for protein precipitation but also for other ion specific effects on proteins, such as crystallization, denaturation, and enzymatic activity.
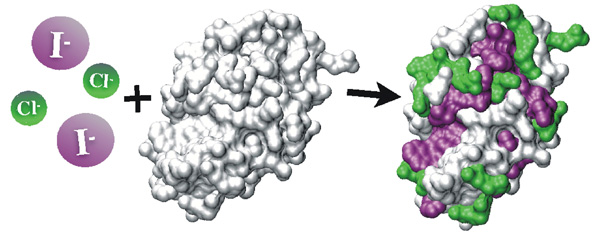
Lund M., Jungwirth P., Woodward C.E.:
Lund M., Vácha R., Jungwirth P:
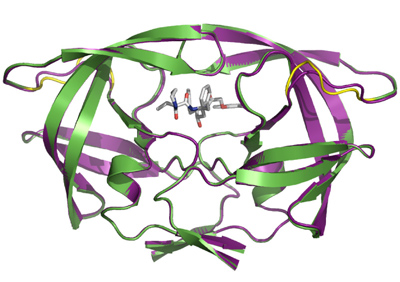
Molecular Characterisation of Drug-Resistant HIV-Protease Mutants with Insertions in the Flap Region Yielding Resistance to Protease Inhibitors.Inhibitors of viral protease are often and successfully used drugs against AIDS. An important complication of successful treatment is the development of virus resistance. Under the selection pressure of inhibitors new virus mutants appear very quickly that are not sensitive to corresponding compounds any more. Most mutations in HIV protease which lead to resistence development are caused by the change of one or more amino acids close to the binding site of the inhibitor. Recently, a novel type of mutations has been described which is based on insertion one or more amino acids into the sequence of viral protease. No corresponding enzyme has been cloned yet, and neither resistance nor its mechanism on a molecular level has been characterized. Together with collleagues from the University Medical Centre in Utrecht we cloned two proteases containing amino acid insertions identified in a Duch patient who underwent long-term treatment by protease inhibitors. We prepared those enzymes by recombinant expression in E. coli and fully characterized them enzymologically. Together with Pavlína Řezáčová team at IOCB we solved their structure by X-ray difraction and suggested mechanism how amino acids insertions can lead to the resistance development against inhibitors of HIV protease. This is the first work of such kind in the literature.
|