Projects
Expression, cellular localization, structure, function and inhibition of glutamate carboxypeptidase II, a neuropeptidase and cancer marker |
|
|
Glutamate carboxypeptidase II is a membrane-bound peptidase expressed in a number of tissues such as kidney, prostate and brain. In the human brain this enzyme cleaves a neurotransmitter N-acetyl-aspartyl glutamate NAAG, liberating thus amino acid glutamate, which plays an important role in a number of pathologic conditions, such as brain stroke neuronal damage, diabetic neuropathy, Alzheimer disease etc. The animal experiments confirmed that GCPII indeed is an important pharmaceutical target for the development of new neuroprotective drugs. The prostate form of the enzyme, known as the Prostate Specific Membrane Antigen (PSMA) is an important cancer marker. We expressed the protein in insect cells and enzymologically characterised it. Specifically, we mapped the PR domain(s) necessary and sufficient for the enzyme activity, analysed the glycosylation of the enzyme, elucidated its substrate specificity and determined (in collaboration) the X-ray structure of the enzyme (free and in complex with number of specific inhibitors). Using novel, specific monoclonal antibodies we are analyzing the expression of the enzyme in various human and animal tissues with the aim to elucidate yet unknown, non-proteolytical role of GCPII in cancerogenesis. [relevant papers: Šácha et al. (2007), Mesters et al. (2006), Bařinka et al. (2002)]. |

|
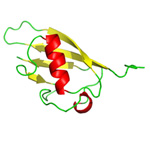
Elucidating the cellular role of DNA damage-inducible protein (Ddi) |
|
|
Our group become also interested in the ubiquitin-proteasome system, mainly one of the ubiquitin-like proteins - Ddi. The yeast ortholog of the DNA damage-inducible protein is considered to play a role as a protein shuttle that transports certain proteins designated for degradation into the proteasome. It harbors three domains: the ubiquitin-like domain (UBL) that interacts with the proteasome, the ubiquitin-associated domain (UBA) that binds ubiquitinated substrates and, quite interestingly, the retroviral protease-like domain (RVP), which is responsible for the dimerization of the whole protein (and which originally attracted our interest due to our long-term commitment to the analysis of retroviral proteins). Although the overall fold of this domain is very similar to retropepsins including highly conserved catalytical triad [DT/SG], the putative proteolytical activity of this domain remains to be clarified. There are two human homologs of the Ddi protein. Surprisingly, their cellular role has not been revealed so far. And that is the ambitious goal of this project: we employ biochemistry, structural biology, genetics, cell biology as well as proteomics to uncover their function. |
|
Structure - function analysis and inhibition of HIV protease developed under the selection pressure in HIV-positive patients treated with protease inhibitors |
|
|
Infection by Human Immunodeficiency virus (HIV) causes estimated 4.9 milion death every year. Therefore, the design of agents blocking HIV replication remains very important challenge. One class of virostatics, inhibitors of HIV proteinase, are especially important. However, high mutation rate of HIV leads to rapid development of resistancy. In our broad-based project, we follow Czech HIV-positive patients treated by protease inhibitors at the University Hospital Bulovka, amplify, sequence and express corresponding recombinant proteases, characterise them enzymologically and structurally. The aim of the research is to analyze the evolution of resistant viral species on molecular level and to identify novel inhibitors, capable of blocking the resistant viral species. [relevant papers: Cígler et al. (2005), Weber et al. (2002)]. |

|
Serine racemase from human brain: an unexpected role for a D-amino acid in the central nervous system |
|
|
Almost all proteins in living organisms are made up from L-amino acids. It has been shown very recently that a D-form of an amino acid, serine, is present in mammalian brains in high concentrations and serves as an important neurotransmitter. D-serine is a co-agonist in the "glycinate site" of N-methyl-D-aspartate receptor (NMDA). These reseptors are in the mammalian central nervous system important for number of physiogical processes including brain development, learning and memory. Overactivation of NMDA receptors might lead to neuronal death. Inhibitors of serine racemase thus might represent possible treatment options for a number of neurodegenerations, including Alzheimer's or Parkinson's diseases. D-serine is formed by serine racemase. We cloned, expressed and characterized recombinant human and mouse serine racemase, developed an activity assay, identified novel substrates and inhibitors of the enzyme and currently are working on the structure-activity analysis of this pharmacologically very relevant protein. [relevant papers: Stříšovský et al. (2005), Stříšovský et al. (2003)]. |

|