2011
|
High resolution structure of a monomeric retoviral protease solved due to game playersAll current inhibitors of HIV proteases that are used in clinical treatment of AIDS are targeted against dimeric form of protease. Retrovirus M-PMV infects rhesus monkeys and causes simian immunodeficiency syndrom (SAIDS). This virus is an excellent model for investigation of many processes in retroviruses and for development of new drugs against retroviruses. Our previous biochemical and NMR studies have indicated that in the absence of substrate or inhibitors M-PMV PR should fold into a stable monomer. For solution of crystal structure we prepared a protease mutant, which enabled to prepare a highly concentrated sample for crystallography. However, crystallographers in Poznan could not solve the structure by available molecular replacement and Rosetta programmes. A solution was finally obtained, after several years of experiments, by players of the online game FoldIt who were able to generate model of sufficient quality for successful molecular replacement and subsequent structure determination. The crystal structure will be used for design of novel types of inhibitors against retroviruses.

to its HIV-1 counterpart extracted from the dimeric context (B). "Flap" marks an important loop, which has a completely unexpected conformation in (A).
Nature Structural and Molecular Biology 18: 1175-1177 (2011).
Gilski M., Kazmierczyk M., Krzywda S., Zábranská H., Cooper S., Popovic Z., Khatíb F., Dímaio F., Thompson J., Baker D., Pichová I., Jaskolski M.:
From inhibition of a digestive enzyme of the human blood fluke to the treatment of schistosomiasisSchistosomiasis (bilharzia) is a chronic infectious disease caused by blood flukes of the genus Schistosoma. It is a global health problem as these helminth parasites infect over 200 million people in tropical and subtropical areas. Treatment and control of schistosomiasis now relies on just one drug, and there is pressure to identify new anti-schistosomal chemotherapeutics. Adult schistosomes live in the cardiovascular system, and host blood proteins are a primary source of nutrients. Cathepsin B1 (SmCB1) is a critical proteolytic enzyme involved in protein digestion in the parasite gut; it has been evaluated as a therapeutic target. We report the first crystallographic structure of SmCB1 and its complexes with peptidomimetic inhibitors (see figure). A comprehensive biochemical analysis is provided that defines relationships among structure, enzymatic activity and inhibition of SmCB1. We directly demonstrate that inhibitors of SmCB1 are toxic to schistosomes. Our results provide a footing for the rational design of inhibitory compounds for the development of novel anti-schistosomal drugs.
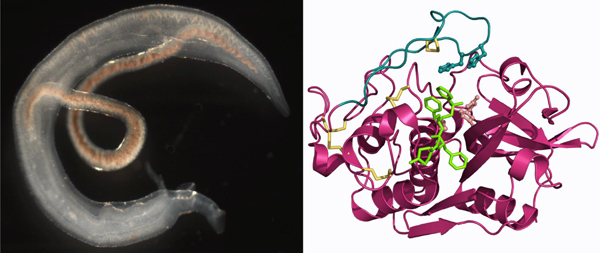
Right: 3D structure of SmCB1 in complex with a synthetic inhibitor.
Jílková A., Řezáčova P., Lepšík M., Horn M., Váchová J., Fanfrlík J., Brynda J., McKerrow J.H., Caffrey C.R., Mareš M.:
Chemical protection of DNA from enzymatic cleavageA very facile and straightforward two-step synthesis of the protected DNA consisting of the synthesis of silyl-protected nucleoside triphosphate followed by enzymatic incorporation to DNA by polymerase was developed. Using PCR, any desired length and sequence of DNA decorated by the bulky protecting groups in the major groove can be prepared. This protected DNA is resistant to cleavage by the restriction endonucleases even if it contains the recognition sequence for the particular enzyme. When this DNA is treated with ammonia, all bulky silyl groups are cleaved off and the resulting DNA modified by small acetylene groups is then recognized and cleaved by the enzymes. It is the first switchable protection of DNA from cleavage and in general also from specific interactions with a protein. Switching of the cleavage is useful in molecular biology for manipulations of large DNA stretches where the recognition sequence might be present is several copies.
 Using PCR, any desired length and sequence of DNA decorated by the bulky protecting groups in the major groove can be prepared. This protected DNA is resistant to cleavage by the restriction endonucleases even if it contains the recognition sequence for the particular enzyme. When this DNA is treated with ammonia, all bulky silyl groups are cleaved off and the resulting DNA modified by small acetylene groups is then recognized and cleaved by the enzymes. It is the first switchable protection of DNA from cleavage and in general also from specific interactions with a protein.
Kielkowski P., Macíčková-Cahová H., Pohl R., Hocek M.: |