|
|
|
Close Help | ||||||||||||||
Our results show the activation of yeast neutral trehalase Nth1 by 14-3-3 proteins from the structural point of view. Our data could be important for understanding of the activation process of Nth1 as well as of the role of 14-3-3 proteins in the regulation of other enzymes.
Our research team has been studying the 14-3-3 proteins which are highly conserved regulatory molecules found in all eukaryotes. First they have been isolated from the bovine brain and their unusual name “14-3-3”, originates from their elution and migration pattern on two-dimensional DEAE-cellulose chromatography and starch gel electrophoresis. 14-3-3 proteins have the ability of binding the functionally different signal proteins, including kinases, fosfatases and transmembrane receptors by changing their function. Through the functional modulation of a wide range of binding partners, 14-3-3 proteins are involved in many processes, including cell cycle regulation, metabolism control, apoptosis,and control of gene transcription.

Structure of the human 14-3-3 protein (isoform zeta) with the modeled C-terminal segment.
Neutral trehalase 1 (Nth1) is an enzyme from the hydrolase family, which takes care of the degradation of non-reducing disaccharide trehalose on two molecules of glucose. Both yeast isoforms of 14-3-3 proteins are responsible for the complete enzyme activity of Nth1 after phosphorylation by PKA in vitro. In our work we identified key phosphorylation sites responsible for the activation of this enzyme (serines on the positions 60 and 83).
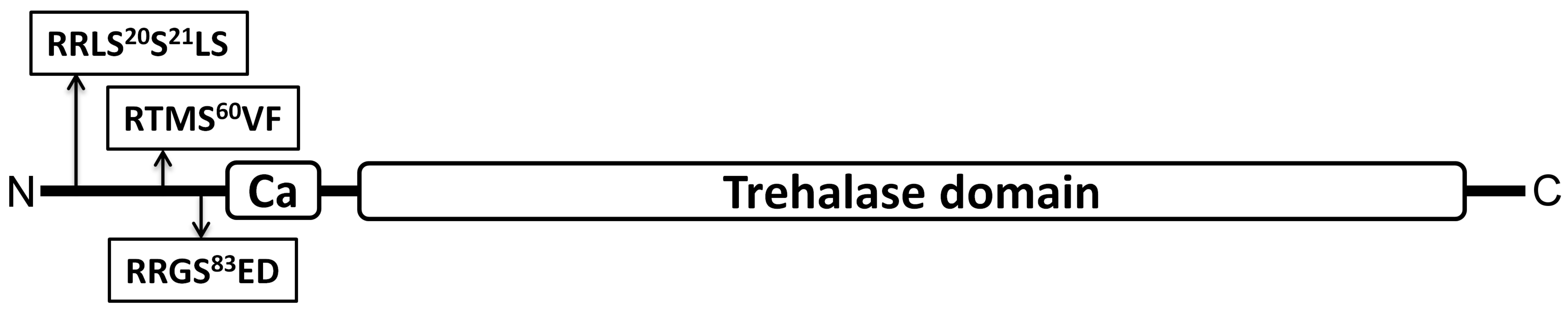
Diagram of S. cerevisiae Nth1 primary structure with the marked phosphorylation sites Ser20, 21, 60 a 83, calcium binding domain and trehalase domain (Veisova et al., 2012).
Yeast 14-3-3 proteins and neutral trehalase Nth1
In 2010 we proved by the series of biochemical and biophysical method that C-terminal segment of yeast 14-3-3 proteins doesn´t function as an autoinhibitor like at other proteins from this family (Veisova et. al., 2010).
In 2012 we have identified key phosphorylation sites responsible for the activation of the enzyme neutral trehalase by the 14-3-3 protein (Veisova et al., 2012).
Recently we are trying to characterize the enzyme neutral trehalase in more detail. We observed, that the 14-3-3 protein binding influence several regions of Nth1 and the formation of complex with 14-3-3 protein is accompanied by the change in tertiary structure (Macakova et al., 2013).
To investigate the structural basis of interaction between Nth1 and Bmh1, we used hydrogen/deuterium exchange coupled to mass spectrometry, circular dichroism spectroscopy and homology modeling to identify structural changes occurring upon the complex formation. The results presented here provide a structural view of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1, which might be relevant to understand the process of Nth1 activity regulation as well as the role of the 14-3-3 proteins in the regulation of other enzymes.

Homology model of the catalytical domain of yeast neutral trehalase regions showing the biggest changes upon the complex formation with 14-3-3 protein are colored in red (Macakova et al., 2013).
Current projects involve: