For genetic dissection of complex pathophysiological traits in recombinant inbred (RI) strains, it is possible to take the advantage of accumulated genotypes and intermediary phenotypes. Intermediate phenotypes have simpler genetic architectures and can be used for connecting variabilty at the DNA level with complex pathophysiological traits. Abundance of mRNA (transcriptome), proteins (proteome), metabolites (metabolome), etc. are the most widely used intermediary phenotypes.
For genetic dissection of complex pathophysiological traits in the SHR, it is possible to take the advantage of accumulated genotypes and intermediary phenotypes in the BXH/HXB recombinant inbred strains.
Recombinant inbred (RI) strains are derived from F2 individuals obtained by crossing of two highly inbred strains. Randomly chosen F2 pairs are brother-sister mated for more than twenty generations to become genetically fixed. Individual RI strains have unique combinations of loci derived by segregation and recombination of the alleles present in the progenitor strains. An important feature of RI strains is that, because they are inbred, repeated assays can be made so that phenotypes of each strain can be more precisely estimated using average results obtained from measurements in multiple rats per each RI strain. A precise estimation of phenotype-genotype relationship is especially important when studying highly variable traits such as blood pressure or behavior. Moreover, because data are cumulative across assays, studies, and research groups, accumulated data can be analyzed and trait relationships studied in a manner not possible with conventional genetically segregating populations. This is a major advantage for the analysis of complex pathophysiological traits. BXH/HXB RI strains were derived from spontaneously hypertensive rat (SHR) and normotensive Brown Norway (BN-Lx) rat.
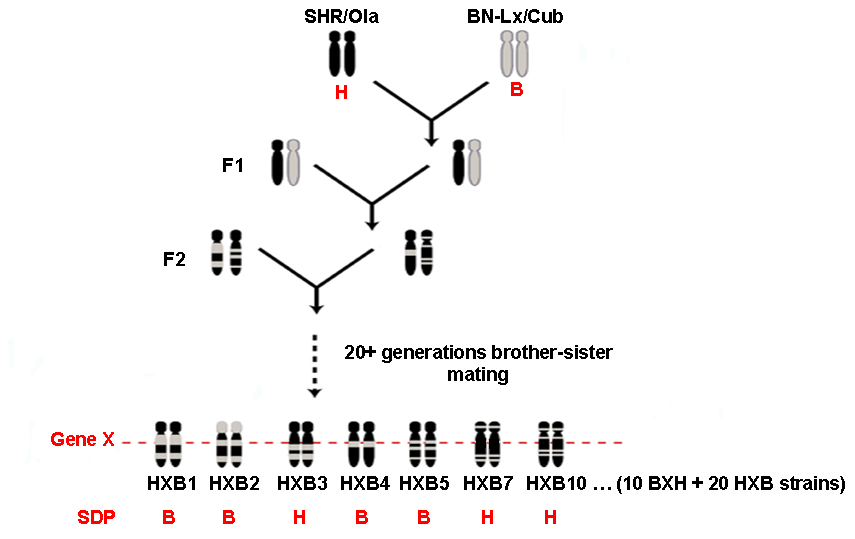
Scheme of derivation of the BXH/HXB recombinant inbred strains
Accumulated data available for linkage and corelation analyses in 30 BXH/HXB RI strains:
Intermediary phenotypes:
Most of the above mentioned intermediary phenotypes, genotypes, and genome sequences were obtained thanks to international collaborative effort within the EURATools and EURATRANS projects of the 6th and 7th Framework of the EC and are available in public databases:
WebQTL (www.genenetworrg.org) and PhenoGene Informatics (http://phenogen.ucdenver.edu/PhenoGen/) databases contain genotypes, gene expression profiles and physiological traits in RI strains. Rat Genome Database (http://rgd.mcw.edu/) contains genome sequences of the SHR/Ola a BN-Lx/Cub progenitors.
Intermediate phenotypes have simpler genetic architectures and can be used for connecting variabilty at the DNA level with complex pathophysiological traits. For instance, abundance of mRNA in tissues is a highly heritable trait and represents very useful intermediary phenotype since it is possible to identify cis- and trans-regulated expression quantitative trait loci (eQTL) as candidate genes for complex traits. The most promising candidate genes are those that are (1) located close to the peak of QTL linkages of pathophysiological traits, (2) whose expression is regulated in cis, and (3) that correlate with the pathophysiological complex traits. The avalability of genome sequences of both progenitor strains, the SHR and BN-Lx, enables rapid screening for functional variants of such candidate genes. We used this approach to identify the first QTL at the molecular level in the SHR, including mutant Ogn (osteoglycin) and Endog (endonuclease G) genes that predispose to left ventricular hypertrophy, deletion variant of Cd36 (fatty acid translocase) that is associated with metabolic disturbances and hypertension or Ebi2 (Epstein-Barr virus induced gene 2) gene that is associated with an inflammatory gene network and its human ortholog with predisposition to Type 1 diabetes. Using similar approach, additional candidate genes have been identified, including genes associated with cardiac microvascular remodeling, catecholamine sythesis and secretion or prefence to alcohol.