Sample Quality and Quantity Requirements
Expression, Exon and Tiling chips - eukaryotic RNA samples
The quality of the total RNA samples should be assessed by agarose gel electrophoresis or Agilent 2100 Bioanalyzer. The total RNA profile should exhibit discrete 28S and 18S ribosomal RNA bands. The 28S rRNA band should be approximately 2 times more intense than the 18S rRNA band. No smear below each rRNA band or shift in RNA size distribution towards smaller fragments should be observed. The A260/A280 ratio should range between 1.8 and 2.0.
To eliminate inhibition of enzymatic reactions during the probe synthesis, the control PCR with fragment of the minimal length of 500bp must be performed on each sample. Alternatively, the PCR with a shorter fragment amplified in the minimal distance of 700bp from the poly(A) end should be performed.
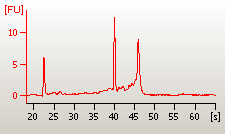
The information proving the quality of RNA samples (either gel images or Bioanalyzer results) must be enclosed to the sample submission form. It is much cheaper to make required quality checks than waste the chips and reagents with poor starting material.
Please, do not resuspend your RNA samples in buffers containing SDS or other detergents as these will inhibit the enzyme reaction used to synthesize the probe. The total RNA must be diluted in deionized water in the minimal concentration for each type of chip and protocol. For the expression chips, one-cycle or two-cycle target labelling protocol is performed depending on the amount of the starting material.
| Chip type (protocol) | Minimal concentration | Required amount of total RNA |
|---|---|---|
| Expression chips (One-Cycle Kit) | 0.3 µg/µl | 6 µg |
| Expression chips (Two-Cycle Kit) | 20 ng/µl | 80 ng |
| Exon chips | 0.5 µg/µl | 3 µg |
| Tiling chips | 0.3 µg/µl | 6 µg |
Expression chips - prokaryotic RNA samples
For Pseudomonas aeruginosa and E. coli chips, total RNA must be free from chromosomal DNA contamination. It is suggested to use the QIAGEN RNeasy Mini Purification Kit and that no more than 10^9 cells are applied to a single purification column. Also, the lysozyme at the concentration of 1 mg/mL and not the recommended 400 µg/mL should be used. Additional DNase I treatment may be required to eliminate DNA contamination when the bacterial culture is grown at high density.
The total RNA must be diluted in deionized water in the minimal concentration. The A260/A280 ratio should be approximately 2.0 with ranges between 1.8 to 2.1 considered acceptable. On an agarose gel the 23S and 16S rRNA bands should be clear without any obvious smears and high molecular weight bands.
| Chip type | Minimal concentration | Required amount of total RNA |
|---|---|---|
| Expression chips (prokaryotic) | 0.7 µg/µl | 15 µg |
Genotyping chips - DNA samples
High-quality genomic DNA is used as a starting material. The DNA has to be double-stranded and without any signs of high degradation. The DNA must be also free from PCR inhibitors and contaminants from other genomic DNA sources (either human or other origin).
The DNA must be diluted to the required concentration in reduced EDTA TE buffer (0.1 mM EDTA, 10 mM Tris HCl, pH = 8.0) since an elevated EDTA level may interfere with subsequent reactions. The A260/A280 ratio should be approximately 2.0 with ranges between 1.8 to 2.1 considered acceptable.
| Chip type | Required concentration | Required amount of genomic DNA |
|---|---|---|
| Mapping 10K chips | 50 ng/µl | 500 ng |
| Mapping 100K and 500K chips | 50 ng/µl | 1 µg |