The study of structure and function of 14-3-3 proteins and their complexes
Our research team has been studying the 14-3-3 proteins which are highly conserved regulatory molecules found in all eukaryotes. 14-3-3 proteins have the ability of binding the functionally different signal proteins, including kinases, fosfatases and transmembrane receptors by changing their function. Through the functional modulation of a wide range of binding partners, 14-3-3 proteins are involved in many processes, including cell cycle regulation, metabolism control, apoptosis, and control of gene transcription. More than 300 proteins have been described as binding partners till now. We employ both biophysical (fluorescence spectroscopy, analytical ultracentrifugation, SAXS, mass spectrometry, isothermal titration calorimetry, X-ray crystallography, protein structure modeling, etc.) and biochemical (recombinant protein expression, site-directed mutagenesis, enzyme kinetics) approaches to understand the details of how the activity and function of protein-protein complexes are regulated.
Research Projects
The 14-3-3 proteins are a family of regulatory molecules, which specifically bind to phosphoserine (or phosphothreonine)- containing motifs (pSer/pThr) in a sequence-specific manner. Through these binding reactions, the 14-3-3 proteins play key regulatory roles in signal transduction, cell cycle control, metabolism control and apoptosis. More than 200 14-3-3 binding partners have been reported so far and some of them play prominent roles in cancer development (e.g. transcription factors p53 and FOXO), neurodegeneration (e.g. Tau protein, ASK1 kinase), cardiovascular diseases (e.g. RGS proteins, phosducin) or inflammation (e.g. NFkB, ASK1 kinase). However, the detailed mechanisms of the14-3-3 protein-mediated regulations are mostly elusive, mainly due to the lack of structural data.
Main goal of our research is a mechanistic understanding of the 14-3-3 protein function in the regulation of selected 14-3-3 protein binding partners. In recent years we have been studying the 14-3-3 protein-mediated regulation of forkhead transcription factor FOXO4, tyrosine hydroxylase, or regulator of G-protein signaling RGS3. Our current projects are focused on regulation of caspase-2, proten kinase CaMKK2, FOXO-DBD, neutral trehalase Nth1, protein kinase ASK1.
Caspase-2
Caspase-2 (C2), a cysteine-dependent and aspartate-specific intracellular protease, has multiple roles in the DNA damage response, cell cycle regulation and tumor suppression. C2 functions as a central coordinator between the cell metabolism and apoptosis and its function is regulated by phosphorylation at several Ser residues. Phosphorylated procaspase-2 (proC2) binds to the 14-3-3 protein and this interaction blocks proC2 activation through an unknown mechanism. To elucidate this regulatory mechanism we propose to: i) identify sites responsible for the 14-3-3 protein binding to proC2, ii) perform biophysical characterization of the 14-3-3:proC2 complex using analytical ultracentrifugation, iii) map the binding interface of the 14-3-3:proC2 complex and perform its structural analysis using hydrogen-deuterium exchange coupled to MS, SAXS and protein crystallography. The proposed research is expected to provide the structural insight into the 14-3-3-dependent regulation of C2. This project is funded by Czech Science Foundation (Project 17-00726S).
CaMKK2
The Ca2+/calmodulin-dependent protein kinase (CaMK) cascade is involved in the regulation of many physiological and pathophysiological processes. This signaling cascade consists of CaMKI and CaMKIV and their upstream activator CaMK kinase (CaMKK). The activity of CaMKK is inhibited through phosphorylation by PKA in a process involving the binding to the 14-3-3 protein. However, the molecular mechanism of this 14-3-3-mediated inhibition of CaMKK is currently unknown. Anticipated mechanisms include direct inhibition through structural modulation of the catalytic site, blocking of dephosphorylation of inhibitory phosphorylation sites or interference with the binding of Ca2+/calmodulin to CaMKK. Main goal of this project is to elucidate the molecular basis of this regulation by performing functional and structural analysis of interactions between 14-3-3 and CaMKK2. This project is funded by Czech Science Foundation (Project 16-02739S).
FOXO3-DBD
FOXO transcription factors control apoptosis, stress resistance and longevity in mammalian cells. Although the members of the FOXO family act as tumor suppressors in some cell types, emerging evidence suggests that FOXO3 also contributes to tumor stem cell renewal, immune suppression, metastases and chemotherapy resistance in certain cancer types. By a combined in silico / cell biological screening approach several small, drug-like compounds that interact with the DNA-binding domain of FOXO3 and efficiently inhibit its transcriptional activity have been identified. The main aim of this project is to define the structural basis for the interaction between compounds and FOXO3 by NMR spectroscopy, design and develop derivatives with improved properties regarding solubility and affinity, analyze compound-dependent inhibition of target recognition by FOXO3 in vitro and in vivo, study the effects of compounds on FOXO3-induced cancer cell survival in 3D cell culture and in vivo. This project is a collaboration with the group of prof. Michael J. Ausserlechner from Medical University Innsbruck, Innsbruck, Austria, and it is funded by Czech Science Foundation (Project 17-33854L).
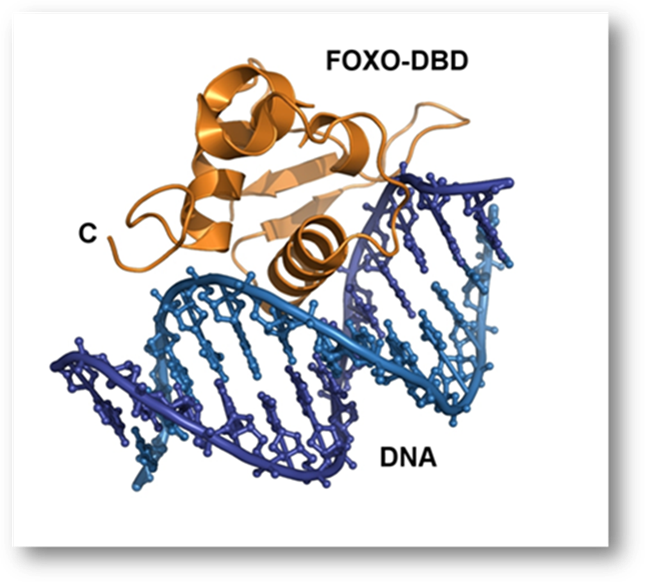
Crystal structure of the FOXO4-DNA-binding domain bound to DNA (PDB 3L2C) published in Boura et. al., Acta Crystallographica D 2010, 66, 1351-7:
Neutral trehalase Nth1
The overall goal of this project is to understand the structural basis of BMH–dependent regulation of the activity ofthe enzyme Neutral trehalase (Nth1). Site-directed mutagenesis, steady-statefluorescence spectroscopy, time-resolved fluorescence spectroscopy, Xray crystallography, analytical ultracentrifugation and enzyme kineticsmeasurements will be the principal tools. Nth1 is responsible for trehalosedegradation and is required in a variety stress conditions. Activity of the Nth1 enzyme was just recently found to be mediated by Bmh1 and Bmh2 binding inyeast. The role of yeast Bmh proteins in the regulation of Nth1 seems toincreasethe enzymatic activity of the Nth1 enzyme. However details concerning interaction between the Bmh proteins and Nth1 remain still unknown. Sincetrehalose metabolism is an essential component of the stress response inyeastcells, thus, the elucidation of the mechanisms of Nth1 protein regulation,which is still unresolved, would provide us with important information concerning mechanisms of both the 14-3-3 protein function and the regulation of the neutral trehalase activity in yeast. This project was funded by Czech Science Foundation (Project P207/11/0455).
Results were published in: Veisova et. al., Biochemical Journal 2012, 443:663-670; Macakova et. al., BBA-Gen. Subjects 2013, 1830:4491-4499; Kopecka et. al., JBC 2014, 289:13948-13961
Specific trehalase activity of pNth1 WT and mutants upon the Bmh1 activation in vitro.
SAXS low-resolution structure of the pNth1:Bmh1 complex.
Proposed mechanism for the 14-3-3 protein-dependent activation of Nth1. The active site (AS) of Nth1 in the absence of Bmh (yeast 14-3-3 isoform) is buried within the structure of the trehalase domain and the enzyme is catalytically inactive. Phosphorylated Nth1 is recognized by Bmh protein and its binding to the N-terminal pSer60 and pSer83 induces conformational change within both the Ca2+-binding domain (shown in orange) and the catalytic domain in close proximity to the active site. This structural change enables substrate and product entry and departure, respectively, hence the enzyme activation.
Protein kinase ASK1
ASK1 is a member of the mitogen-activated protein kinase kinase kinase family, activates JNK and p38 MAP kinase signaling pathways in response to various stress stimuli, including oxidative stress, endoplasmic reticulum stress, and calcium ion influx . ASK1 plays a key role in the pathogenesis of multiple diseases including cancer, neurodegeneration and cardiovascular diseases and is considered as a promising therapeutic target. The activity of ASK1 is regulated by several other proteins including thioredoxin and the 14-3-3 protein that both function as physiological inhibitors of ASK1. The main goal of this project is to study the role of the 14-3-3 proteins in the regulation of the kinase activity of the catalytic domain of protein kinase ASK1 (Apoptosis signal-regulating kinase 1). The 14-3-3 protein binding to the catalytic domain of ASK1 inhibits ASK1 function through unknown mechanism. To better understand this process we propose: (i) to prepare protein complexes between the catalytic kinase domain of ASK1 and various 14-3-3 protein isoforms; (ii) to determine enzyme kinetic parameters of the kinase domain of ASK1 both in its apo state and in complex with various 14-3-3 protein isoforms; (iii) to map protein-protein interactions within the prepared complexes and to perform their biophysical and structural characterization. Recombinant protein expression, site-directed mutagenesis, methods of enzyme kinetics, analytical ultracentrifugation, mass spectrometry, fluorescence spectroscopy, protein crystallography and SAXS will be used as principal tools. This project was funded by Czech Science Foundation (Project 14-10061S).
Results were published in: Kosek et al. (2014) J. Biol. Chem.; Petrvalska et al. (2016) J. Biol. Chem.; Kylarova et al. (2016) FEBS J.
Phosducin (Pdc)
Pdc is a highly conserved acidic phosphoprotein that regulates visual signal transduction by modulating the amount of transducin Gtαβγ heterotrimer through competitionwith the Gta subunit for binding to the Gtβγ complex. Pdc was also shown to regulate the cardiovascular system by modulating sympathetic activity and blood pressure. Phosducin function is regulated through the phosphorylation and the binding to the 14-3-3 protein. This project was funded by Czech Science Foundation (Project P305/11/0708).
Results were published in: Rezabkova et al. (2012) Biophys. J.; Kacirova et al. (2015) J. Biol. Chem.; Kacirova et al. (2017) Biophys. J.